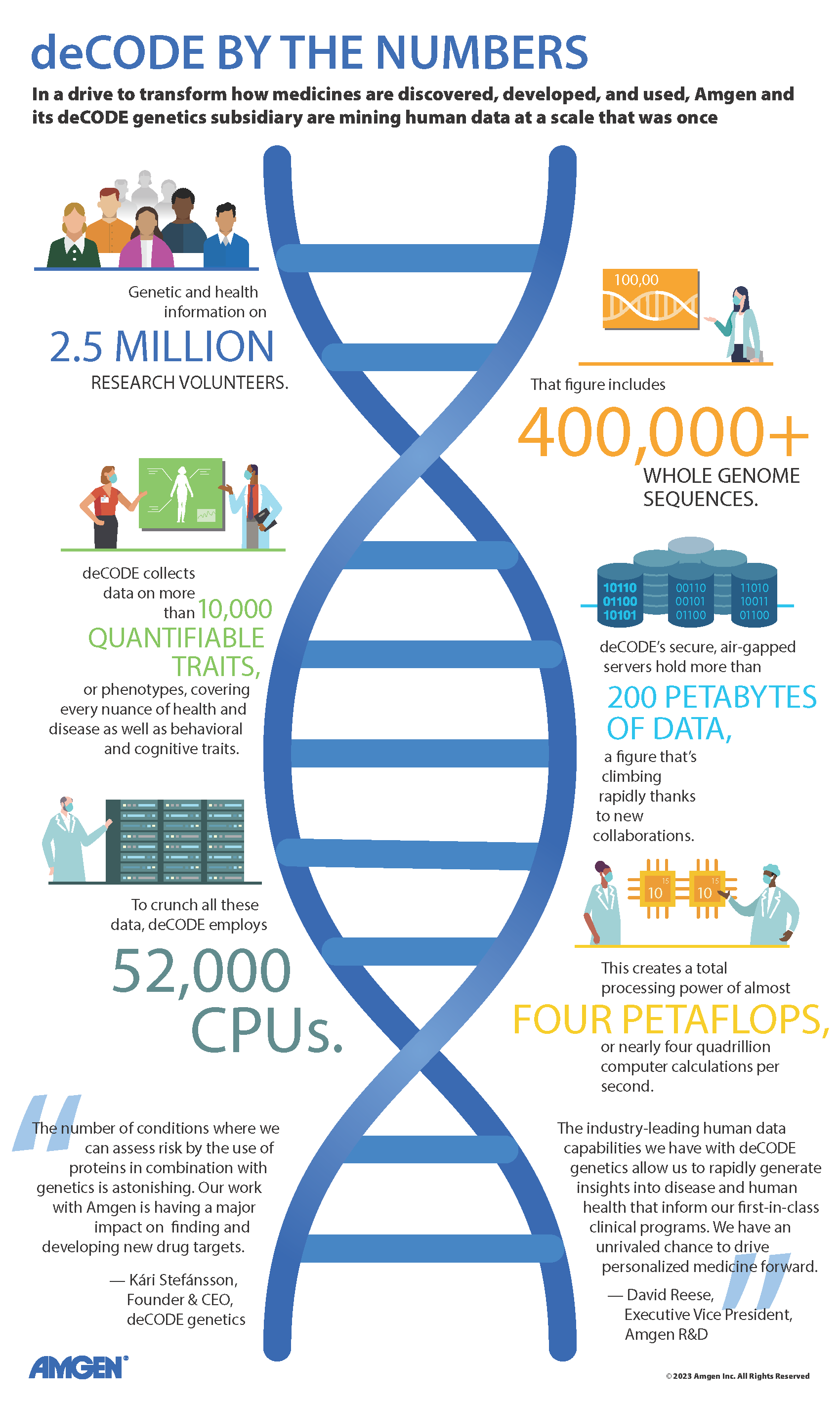
When the human genome was first fully sequenced, it was hailed as an overwhelming success. Since that time there has been an enormous amount researchers have learned from the information contained in the diversity of the 3 billion base pairs that make up our genome.
But before the human genome sequence was even complete, a small Icelandic company was busy trying to understand human diversity by taking a population-scale approach to studying the human genome.
Since its founding in 1996, deCODE genetics has contributed a tremendous amount to the research community’s understanding of human genetic variation. Seeing the value of using population genomic research to understand diversity as it applies to disease was a big draw for Amgen, who acquired deCODE in 2012.
A new way to study human diversity and biology
As deCODE joined Amgen, large-scale genome sequencing was taking off. The idea of studying the human genome to better understand the biochemical pathways involved in disease provided a much better model than the current standard of studying these pathways in mice.
“The problem with mice is they aren’t terribly good at predicting what will happen in humans,” said Bob Bradway, Amgen’s CEO. “That’s one reason why it takes 10 to 15 years and costs $2.5 billion to advance a drug from the earliest stages to approval.” Amgen researchers reasoned that studying diseases in large populations rather than in preclinical models might increase the success of drug discovery and development, said Bradway. He added that a substantial portion of Amgen’s research and development pipeline is now supported by genetic insights from deCODE.

deCODE has been at the forefront of adopting new technologies, such as whole genome sequencing, and implementing them at scale. deCODE uses high-throughput sequencers (shown here) that produce both short and long read sequences. Long reads are helpful in determining structural variations, including parts of the genome that are deleted, inserted, repeated, duplicated or inverted. Short reads are less error prone.
Looking at the entire genome using Whole Genome Sequencing (WGS) allows researchers to search for changes in any part of the genome in a completely bias-free way. This means the researchers follow the data, rather than making a hypothesis that could narrow the focus to only certain areas of interest in the genome. “There is hardly anything more dangerous to a scientist than a great hypothesis,” said deCODE genetics founder, Kári Stefánsson. “You have to be humble and accept what the data give you.”
Genome wide association studies (GWAS) look for common variations in DNA’s letters, or single nucleotide polymorphisms (SNPs), across the genome. The studies then use these data to find associations between common SNP variants and diseases or other traits. Over the past decade GWAS studies have helped deCODE identify thousands of sequence variants that are associated with a whole host of traits, including BMI, height, and conditions such as cardiovascular, neurodegenerative and inflammatory diseases.
For example, the deCODE team discovered a variant in the gene TREM2 (triggering receptor expressed on myeloid cells 2), that codes for a receptor on microglial cells, which are immune cells called histiocytes that are found in the brain. The variant in TREM2 is associated with an increased risk of Alzheimer’s disease (AD). Subsequent research by deCODE suggests the variant decreases the function of TREM2. That may prevent the microglial cells from removing amyloid deposits from the brain, contributing to the pathogenesis of AD. Amyloid is a well-known protein that has been shown to accumulate and disrupt function in the brains of individuals with AD.
deCODE’s GWAS research has shown that there are many SNP variants associated with common diseases. WGS, which is a more comprehensive way of searching for variants in the genome, has shown rare variants can also associate with common diseases. While rare variants tend to show up within genes, WGS also shows there are many important ones between genes that can still have a significant impact on disease.
Population-scale studies let the data speak louder
Iceland has about 376,000 inhabitants, as well as a single-payer health care system where all phenotypic information is linked to a unique identifier. The country also has a founder effect, which occurs when a population is descended from a small number of ancestors, which reduces genetic diversity. This, along with the nation’s interest in genealogy, makes it an ideal population in which to begin to study human diversity. To date, close to 200,000 Icelanders have participated in one or more of deCODE’s research projects, contributing biological samples, health and medical information and informed consent, showing there is strong support for this kind of research from the larger community.

Blood samples of Icelanders who have participated in deCODE research studies are stored on site. deCODE also runs a phenotyping center that provides clinical assessments of patients to enhance the breadth and depth of their data collection.
Aside from Iceland, deCODE also collaborates with researchers and collects data from populations in the U.S., Europe and Japan. It’s important to have a large enough group of participants to study, since the more people you have, the more power you have to detect associations in genetic research, particularly when looking for rare variants in the genome. “deCODE has helped us move into a position to let the science tell us about the biology with the scale to let the data speak loudly enough,” said Sir Rory Collins, chief scientific officer of the UK Biobank and deCODE collaborator.
The UK Biobank began in 2006 and aims to study how genetics and the environment impact disease. The long-term research study provides access to medical and genetic data from 500,000 participants to accredited researchers around the world. deCODE, along with the Wellcome Sanger Institute, are providing whole genome sequencing for the project. Amgen is one of four corporate partners that is helping to fund the project. Amgen is also collaborating with Intermountain Healthcare, a Utah-based healthcare delivery network, and seeks to do WGS and genotype, or look for SNPs, in 500,000 individuals. “We are working to establish collaborations,” said Stefánsson. “So we collect the data from people of various backgrounds and ethnicities that we so desperately need to find insights and make discoveries that will impact the broadest range of patients and diseases.”
While there are now biobanks in many parts of the world, deCODE was a pioneer in collecting population-scale human data to understand human diversity and disease. The deCODE researchers collect participants’ blood samples to sequence. They pair this with disease data from medical records, registries and questionnaires, as well as quantitative traits such as imaging scans, lipid levels and other test results.
The challenges of data analysis
With so many samples, deCODE has also had to find a way to efficiently analyze all this data. The group has developed sophisticated computational methods that include artificial intelligence or machine learning to mine all this information. When you have such large datasets, computational analyses can find many correlations, but not all of them will be true. That is the challenge faced by groups like deCODE. “I believe the analytical engine that has been built here at deCODE exists in few other places in the world,” said David Reese, Amgen’s executive vice president for Research and Development.
During analyses, a common question is whether an environmental factor causes a disease or is simply part of the course of the disease. One way deCODE researchers are looking at this is through Mendelian Randomization (MR), a technique that assumes that genetic variants that affect an environmental factor should also lead to a proportionally similar increased risk of disease.
For example, researchers at deCODE and colleagues looked at individuals with a variant in the endothelial lipase gene, LIPG, that is linked to having higher levels of the “good” form of cholesterol, HDL. LDL, or the “bad” form of cholesterol, and HDL are among the most commonly measured biomarkers for cardiovascular disease. Elevated levels of HDL are associated with a reduced risk of myocardial infarction (MI), or heart attack. But it has not been clear if the HDL is a direct cause of this reduced risk. Using MR, the research team looked at the effects of the variant on MI risk and found no association. This led them to conclude that raising HDL cholesterol might not reduce risk of MI, challenging a commonly held assumption in medicine. It also emphasizes the limitation of using HDL cholesterol levels as a measure of MI risk.
Layering on multi-omics data
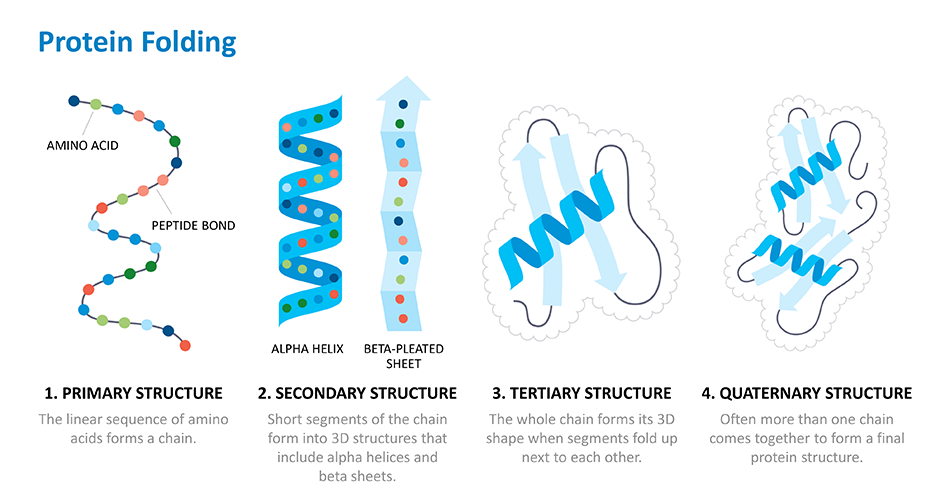
While looking at genomics has served deCODE well, there is also valuable information in other “multi-omics” data. These data can come from transcriptomics, or the study of RNA transcripts made by genes in our DNA, and proteomics, or the study of proteins that ultimately result from those transcripts. Taking a multi-omics approach allows researchers to identify, measure and understand all the biological molecules involved in health and disease.
The levels of proteins in our blood can document both the body’s biological processes as well as the impact the environment has on us. In collaboration with the UK Biobank, Amgen and deCODE are participating in one of the largest proteomic studies measuring circulating proteins to provide further insights into health and disease.
By looking at protein levels in populations with specific diseases, researchers can capture a protein risk score for that disease. The protein risk score changes over time and is likely documenting a disease process that is already progressing. Another commonly used disease risk score is the polygenic risk score, which looks at the effects of multiple sequence variants in the germline DNA that you are born with. Unlike the proteomic risk score, the polygenic score is set at birth and does not change. But the disadvantage of it is that it is most useful earlier on in life, when it can identify risk before an individual gets the disease. deCODE research has shown that polygenic risk scores were not as good as protein risk scores at predicting future disease events when calculated on a group of older individuals who have not been diagnosed with cardiovascular disease.

From left, Rory Collins (UK Biobank), Kári Stefánsson (deCODE genetics), Ray Deshaies (Amgen) and David Reese (Amgen), at a media event at the deCODE headquarters in Reykjavik, Iceland.
Traditionally researchers have used multi-omics data for drug target identification and validation in drug discovery. But only about 20% of the targets identified are druggable. That is where multispecific medicines can close the gap. While researchers may not be able to target 80% of the disease-causing proteins, cells can control every protein in the body. “Cells have figured out how to drug everything in the genome. So we take a page from nature to go after these targets,” said Ray Deshaies, Amgen’s senior vice president of Global Research. Multispecific drugs tap into these existing mechanisms and link them to target proteins involved in disease.
It is also becoming clear that multi-omics data will be highly valuable in drug development. As we age, certain protein levels change and seem to become better predictors of a disease’s progression, rather than contributing to the onset of disease. Drug developers are looking to take advantage of that knowledge to find biomarkers that are associated with individuals whose conditions are progressing quickly and could benefit most from a drug. These biomarkers could help stratify trial populations, provide more targeted treatments and ultimately save time and resources when developing drugs.
“It’s the infusion of these multi-omics techniques into the clinical trials portion of drug development that will be most illuminating, ultimately leading us toward precision medicine,” said Reese. “As deCODE genetics celebrates 25 years, to me, this isn’t an ending, but just a beginning.”